Test Ansible role with Molecule on Macos#
Introduction#
We will go through the steps of creating a molecule test for your role. This will test a roll for CentOS7 on MacOS.
Setup#
Install Docker on your mac#
softwareupdate --install-rosetta
brew install --cask docker
open /Applications/Docker.app
Upload your role to Galaxy#
Note
The address for Galaxy is:
Click on "Login" and then click on the Github icon
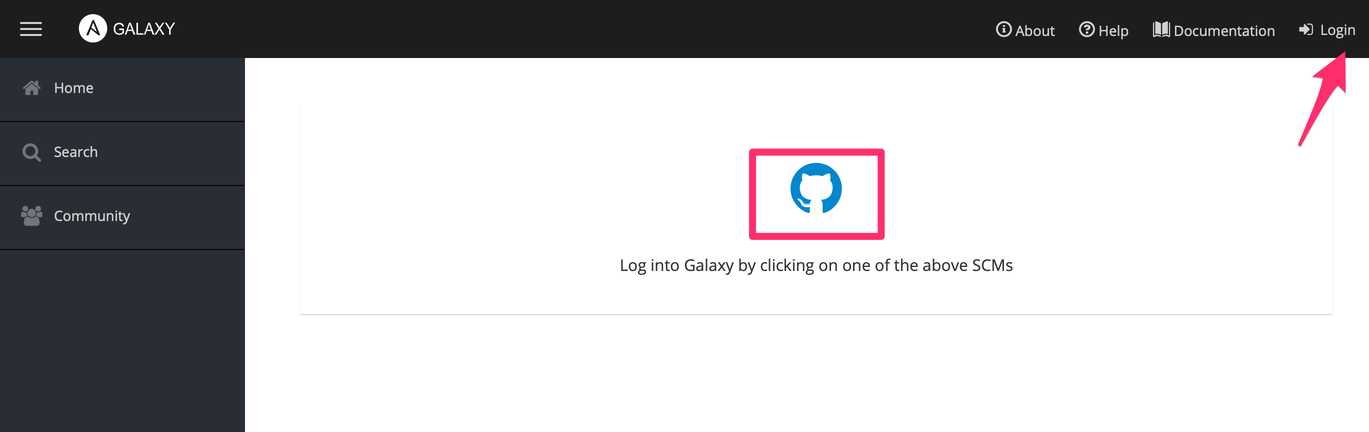
Click on "My Content"
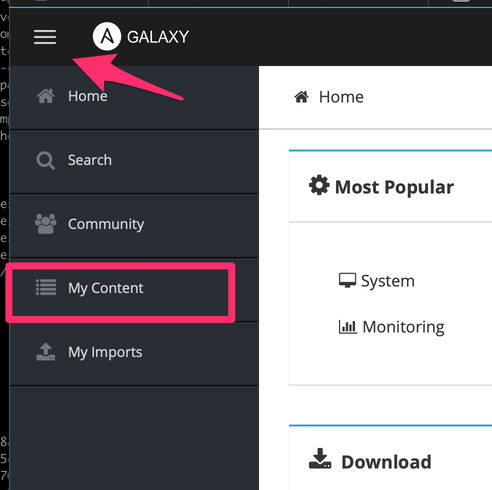
Click on "Add Content"
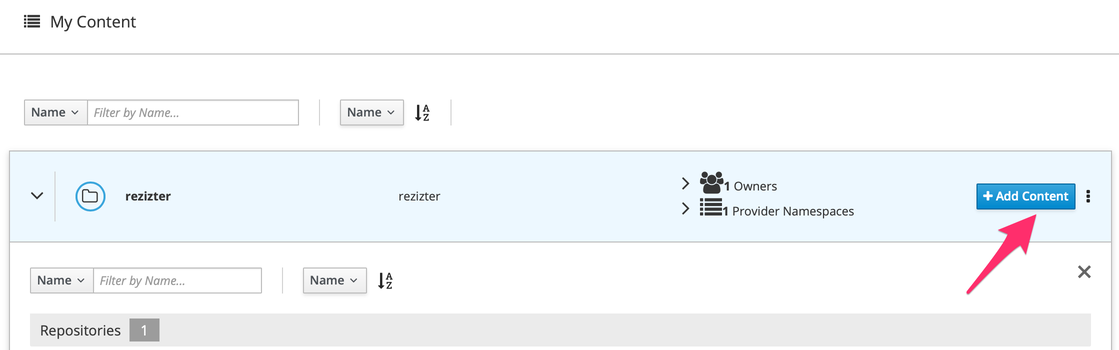
Click on "Import Role from GitHub"
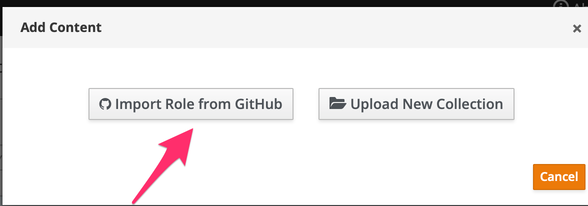
Click on the role you want to import and click on "Ok"
Setup the Molecule role#
Note
The
molecule init role --driver-name docker <ROLE_NAME>
cp -r your_repo/molecule <ROLE_NAME>/
cp -r your_repo/tests <ROLE_NAME>/
Now create the molecule file:
mkdir -p molecule/default
vi molecule/default/molecule.yml
Add:
---
dependency:
name: galaxy
driver:
name: docker
platforms:
- name: instance
image: geerlingguy/docker-centos8-ansible:latest
command: /sbin/init
volumes:
- /sys/fs/cgroup:/sys/fs/cgroup:rw
cgroupns_mode: host
privileged: true
pre_build_image: true
Create a meta file which references your role:
meta/main.yml
Add:
galaxy_info:
role_name: <ROLE_NAME>
description:
license: <YOUR LICENSE>
min_ansible_version: 2.8
platforms:
- name: el
versions:
- 7
- 8
galaxy_tags:
- logging
dependencies: []
Now create the converge file:
vi molecule/default/converge.yml
Add:
---
- name: Converge
hosts: all
tasks:
- name: "Include <ROLE_NAME>"
include_role:
name: "<ROLE_NAME>"
Create a verify file
vi molecule/default/verify.yml
Add:
---
# This is an example playbook to execute Ansible tests.
- name: Verify
hosts: all
gather_facts: false
tasks:
- name: Example assertion
ansible.builtin.assert:
that: true
Create a github directory
mkdir -p .github/workflows
vi .github/workflows/ci.yml
Add
---
name: CI
'on':
push:
branches:
- master
schedule:
- cron: "30 8 * * 0"
defaults:
run:
working-directory: '<ROLE_NAME>'
jobs:
lint:
name: Lint
runs-on: ubuntu-latest
steps:
- name: Check out the codebase.
uses: actions/checkout@v3
with:
path: '<ROLE_NAME>'
- name: Set up Python 3.
uses: actions/setup-python@v3
with:
python-version: '3.x'
- name: Install test dependencies.
run: pip3 install yamllint
- name: Lint code.
run: |
yamllint .
molecule:
name: Molecule
runs-on: ubuntu-latest
strategy:
matrix:
include:
- distro: centos-8
playbook: converge.yml
steps:
- name: Check out the codebase.
uses: actions/checkout@v3
with:
path: 'rezizter.ansible_fqdn'
- name: Set up Python 3.
uses: actions/setup-python@v3
with:
python-version: '3.10'
- name: Install test dependencies.
run: pip3 install ansible molecule molecule-plugins[docker] docker
- name: Run Molecule tests.
run: molecule test
env:
PY_COLORS: '1'
ANSIBLE_FORCE_COLOR: '1'
MOLECULE_DISTRO: ${{ matrix.distro }}
MOLECULE_PLAYBOOK: ${{ matrix.playbook }}
Yamllint#
This is a tool to check the linting of your code before you push to Github
yamllint .
Push the code to Github#
git add -A
git commit -m "add ci and release"
git push
Test using Molecule#
Note
Docker needs to be running locally on your machine to test
Run the following command to test Converge
molecule converge
Github Action#
Once you have pushed to github in the step above, You will see the action testing your code on a CentOs8 docker image.
Go to your repo on GitHub and click on "Actions"
Notes#
Set Cgroup Version to 1#
sed -i '' -e 's/"deprecatedCgroupv1": false/"deprecatedCgroupv1": true/g' Library/Group\ Containers/group.com.docker/settings.json
Restart Docker
killall Docker
open /Applications/Docker.app